Introduction
This article describes the steps needed to create, and analyse, patient data selections from the Discovery database.
Datasets are needed to add patient data to a report. Each dataset represents the results gained from running a combination of SQL commands and stored procedures on a data source.
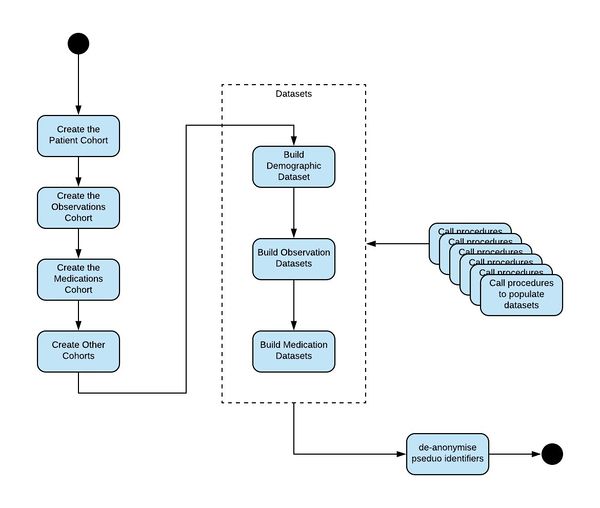
The following diagram shows the algorithm that is followed to generate these datasets:
Patient cohort
Creating the patient cohort
The first step, when a list of patients marked for observational study is received, is to create a patient cohort. Patient data usually comes into the DDS in the form of pseudonymised personal data. Additional information about the patient can be obtained from the Discovery database using the pseudo identifier or the pseudo NHS identifier.
The following entity diagram shows the relationships between patient and organisation:
Patients are linked to the Discovery database using pseudo patient identifiers. However, if only pseudo NHS numbers are provided, the corresponding pseudo identifiers can be derived from the link distributor table. Patients are filtered by organisation ODS codes to ensure that only the patients of the required GP practices are selected.
Building the patient cohort SQL with pseudo NHS numbers
The following example SQL shows how a patient cohort is built using pseudo NHS numbers:
CREATE TABLE cohort_gh2 AS
SELECT
p.id AS patient_id,
p.pseudo_id AS group_by,
p.person_id AS person_id,
p.organization_id AS organization_id,
n.pseudo_nhsnumber AS pseudo_nhsnumber,
p.age_years
FROM gh2_pseudonhsnumbers n
JOIN enterprise_pseudo.link_distributor l
ON n.pseudo_nhsnumber = l.target_skid AND l.target_salt_key_name = 'EGH'
JOIN enterprise_pseudo.patient p ON p.pseudo_id = l.source_skid
JOIN enterprise_pseudo.organization org ON p.organization_id = org.id
WHERE EXISTS
(SELECT 'x' FROM gh2ccg_codes gh2ccgs
WHERE gh2ccgs.parent IN ('City & Hackney CCG','Newham CCG','Tower Hamlets CCG','Waltham Forest CCG')
AND gh2ccgs.local_id = org.ods_code);
Sometimes, the cohort is created with filtered observations.
The example below filters patients who are registered, alive, and have had diabetes. They belong to the age 12 years and over. Two additional tables are introduced for the query: observation(patients who have had diabetes) and episode of care (patients who are registered and have the appropriate registration type for this study).
Patients can be selected with a specific set of observations using a lookup table of relevant SNOMED codes.
Some patients prefer to withhold consent and are therefore are excluded from the cohort; the relevant SNOMED consent code can be checked in the observation table.
Patients are usually only select if they have registration type 2 (Regular/GMS).
Sometimes patients are also based on their age in years.
Building patient cohort SQL with filtered observations
CREATE TABLE cohort_el AS
SELECT DISTINCT
p.id AS patient_id,
p.person_id AS person_id,
p.pseudo_id AS group_by,
org.id AS organization_id,
org.ods_code AS practice_code,
org.name AS practice_name,
e.date_registered AS registered_date,
p.date_of_death AS date_of_death
FROM enterprise_pseudo.observation o
JOIN enterprise_pseudo.organization org ON org.id = o.organization_id
JOIN enterprise_pseudo.patient p ON p.id = o.patient_id
JOIN enterprise_pseudo.episode_of_care e ON e.patient_id = o.patient_id
WHERE EXISTS
(SELECT 'x'FROM data_extracts.snomed_codes s
WHERE s.group_id = 1 AND s.snomed_id = o.snomed_concept_id)
AND NOT EXISTS
(SELECT 'x'FROM data_extracts.snomed_codes s
WHERE s.group_id = 2 AND s.snomed_id = o.snomed_concept_id)
AND org.ods_code IN
(SELECT el.local_id FROM data_extracts.elccg_codes el
WHERE el.parent IN ('City & Hackney CCG','Newham CCG',
'Tower Hamlets CCG','Waltham Forest CCG'))
AND p.date_of_death IS NULL
AND e.registration_type_id = 2 AND e.date_registered <= now()
AND (e.date_registered_end > now() or e.date_registered_end IS NULL) AND (p.age_years >= 12);
Observation cohort
Creating the observation cohort
Observation cohort are created to improve query performance; instead of scanning through the entire observation table, a sub-set of the observation table is created, based on the patient cohort.
Building the observation cohort SQL
CREATE TABLE cohort_gh2_observations AS
SELECT DISTINCT
o.id,
o.patient_id,
o.person_id,
cr.group_by,
cr.pivot_date,
o.clinical_effective_date,
o.snomed_concept_id AS original_code,
SUBSTRING(o.original_term, 1, 200) AS original_term,
o.result_value,
o.result_value_units,
cr.age_years
FROM enterprise_pseudo.observation o JOIN cohort_gh2 cr
ON o.person_id = cr.person_id
AND o.organization_id = cr.organization_id
AND o.patient_id = cr.patient_id;
CREATE INDEX gh2_obs_ix ON cohort_gh2_observations(original_code);
CREATE INDEX gh2_obs_grpby_ix ON cohort_gh2_observations(group_by);
Medications cohort
Creating the medications cohort
Medications cohort are created to improve query performance; instead of scanning through the entire observation table, a sub-set of the medication statement table is created, based on the patient cohort.
Building the medication cohort SQL
CREATE TABLE cohort_gh2_medications AS
SELECT DISTINCT
m.id,
m.dmd_id AS original_code,
m.person_id,
m.patient_id,
cr.group_by,
cr.pivot_date,
SUBSTRING(m.original_term, 1, 200) AS original_term,
m.clinical_effective_date,
m.cancellation_date
FROM enterprise_pseudo.medication_statement m JOIN cohort_gh2 cr
ON m.organization_id = cr.organization_id
AND m.person_id = cr.person_id
AND m.patient_id = cr.patient_id;
CREATE INDEX gh2_med_ix ON cohort_gh2_medications(original_code);
CREATE INDEX gh2_med_grpby_ix ON cohort_gh2_medications(group_by);
Other cohorts
Creating other cohorts
This step can apply to any large tables to support improved query performance. For example, the encounter_raw table is a good candidate to create a separate cohort for SQL query; you can query this table if you want information such as the number of face to face encounters, or the number of home visits.
Building the encounter raw cohort SQL
CREATE TABLE cohort_lbhsc_encounter_raw
AS
SELECT r.person_id,
r.patient_id,
r.clinical_effective_date,
SUBSTRING(UPPER(r.fhir_original_term),1,200) fhir_original_term,
r.organization_id
FROM enterprise_pseudo.encounter_raw r
JOIN cohort_lbhsc c ON r.patient_id = c.patient_id;
CREATE INDEX lbhsc_encounter_term_idx
ON cohort_lbhsc_encounter_raw(fhir_original_term);
CREATE INDEX lbhsc_encounter_person_idx
ON cohort_lbhsc_encounter_raw(person_id);
Building datasets
This involves creating a number of tables in the database.
Patient demographic dataset
A typical patient demographic dataset includes the following information:  This information is translated into a 'create table' script, which is then run to create the dataset table in the database.
This information is translated into a 'create table' script, which is then run to create the dataset table in the database.
Create table script for a patient demographic dataset table
CREATE TABLE gh2_demographicsDataset (
ExtractDate DATETIME NULL,
Pseudo_id VARCHAR(255) NULL,
Pseudo_NHSNumber VARCHAR(255) NULL,
Gender VARCHAR(50) NULL,
Age VARCHAR(50) NULL,
DateOfBirth DATE NULL,
EthnicityLCode VARCHAR(50) NULL,
EthnicityLTerm VARCHAR(200) NULL,
BirthCountryLCode VARCHAR(50) NULL,
BirthCountryLTerm VARCHAR(200) NULL,
RegistrationStart DATE NULL,
RegistrationEnd DATE NULL,
IMD2010 VARCHAR(50) NULL,
LSOA2011 VARCHAR(50) NULL,
PracticeODSCode VARCHAR(50) NULL,
PracticeODSName VARCHAR(255) NULL,
CCGName VARCHAR(100) NULL,
YearOfDeath INT(4) NULL,
F2fVisits_Total INT(11) DEFAULT 0,
F2fVisits_1year INT(11) DEFAULT 0,
F2fVisits_5years INT(11) DEFAULT 0
);
ALTER TABLE gh2_demographicsDataset ADD INDEX gh2Demo_pseudoid_idx (pseudo_id);
The dataset is then populated with pseudo identifiers from the patient cohort.
INSERT INTO gh2_demographicsDataset (pseudo_id, extractdate) SELECT DISTINCT
group_by, now() FROM cohort_gh2;
Separate update SQL queries then populate the columns of the dataset table. For example:
DROP TEMPORARY TABLE IF EXISTS qry_age;
CREATE TEMPORARY TABLE qry_age (
row_id INT,
group_by VARCHAR(255),
age_years INT,
lsoa_code VARCHAR(50), PRIMARY KEY(row_id)
) AS
SELECT (@row_no := @row_no + 1) AS row_id,
c.group_by,
p.age_years,
p.lsoa_code
FROM enterprise_pseudo.patient p JOIN cohort_gh2 c
ON p.pseudo_id = c.group_by, (SELECT @row_no := 0) t;
SET @row_id = 0;
WHILE EXISTS
(SELECT row_id from qry_age
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
UPDATE gh2_demographicsDataset d
JOIN qry_age q ON d.pseudo_id = q.group_by
SET d.Age = q.age_years,
d.LSOA2011 = q.lsoa_code
WHERE q.row_id > @row_id AND q.row_id <= @row_id + 1000;
SET @row_id = @row_id + 1000;
END WHILE;
In the following example, the age and LSOA code are derived from the Patient table. The demographic dataset table is updated with those values when there is a match on the pseudo identifier. If the update is large, it might be possible to optimise the update process by using batches; for example to add IMD2010:
DROP TEMPORARY TABLE IF EXISTS qry_imd;
CREATE TEMPORARY TABLE qry_imd (
row_id INT,
pseudo_id VARCHAR(255),
IMD2010 DECIMAL(5,3), PRIMARY KEY(row_id)
) AS
SELECT (@row_no := @row_no + 1) AS row_id,
d.pseudo_id,
lso.imd_score
FROM gh2_demographicsDataset d JOIN enterprise_pseudo.lsoa_lookup lso
ON d.LSOA2011 = lso.lsoa_code, (SELECT @row_no := 0) t
WHERE d.lsoa2011 IS NOT NULL;
SET @row_id = 0;
WHILE EXISTS
(SELECT row_id from qry_imd
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
UPDATE gh2_demographicsDataset d
JOIN qry_imd i ON d.pseudo_id = i.pseudo_id
SET d.IMD2010 = i.imd_score
WHERE i.row_id > @row_id AND i.row_id <= @row_id + 1000;
SET @row_id = @row_id + 1000;
END WHILE;
Add registration start date, registration end date, practice ODS code, practice ODS name:
DROP TEMPORARY TABLE IF EXISTS reg_sort;
DROP TEMPORARY TABLE IF EXISTS qry_reg;
CREATE TEMPORARY TABLE qry_reg AS
SELECT c.group_by,
c.person_id,
e.date_registered,
e.date_registered_end,
o.name,
o.ods_code,
g.parent
FROM cohort_gh2 c
JOIN enterprise_pseudo.episode_of_care e ON e.person_id = c.person_id
JOIN enterprise_pseudo.organization o ON o.id = e.organization_id
JOIN gh2ccg_codes g ON g.local_id = o.ods_code;
CREATE TEMPORARY TABLE reg_sort (
row_id INT,
group_by VARCHAR(255),
person_id BIGINT,
date_registered DATE,
date_regiostered_end DATE,
ode_code VARCHAR(50),
name VARCHAR(255),
parent VARCHAR(100),
rnk INT, PRIMARY KEY(row_id)) AS
SELECT (@row_no := @row_no + 1) AS row_id,
a.group_by,
a.person_id,
a.date_registered,
a.date_registered_end,
a.ods_code,
a.name,
a.parent,
a.rnk
FROM (SELECT q.group_by,
q.person_id,
q.date_registered,
q.date_registered_end,
q.name,
q.ods_code,
q.parent,
@currank := IF(@curperson = q.person_id, @currank + 1, 1) AS rnk,
@curperson := q.person_id AS cur_person
FROM qry_reg q, (SELECT @currank := 0, @curperson := 0) r
ORDER BY q.person_id, q.date_registered DESC ) a, (SELECT @row_no := 0) t
WHERE a.rnk = 1;
SET @row_id = 0;
WHILE EXISTS (SELECT row_id from reg_sort
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
UPDATE gh2_demographicsDataset d
JOIN reg_sort reg ON d.pseudo_id = reg.group_by
SET d.RegistrationStart = reg.date_registered,
d.RegistrationEnd = reg.date_registered_end,
d.PracticeODSCode = reg.ods_code,
d.PracticeODSName = reg.name,
d.CCGName = reg.parent
WHERE reg.row_id > @row_id
AND reg.row_id <= @row_id + 1000;
SET @row_id = @row_id + 1000;
END WHILE;
In the above example, two temporary tables are created; the first to hold the registration data, and the second to select patient records that have the latest registration date. Add the total number of face to face visits:
DROP TEMPORARY TABLE IF EXISTS noVisitsTotal;
CREATE TEMPORARY TABLE noVisitsTotal (
row_id INT,
group_by VARCHAR(255),
visits INT, PRIMARY KEY(row_id) ) AS
SELECT (@row_no := @row_no + 1) AS row_id,
b.group_by,
b.visits
FROM (SELECT cr.group_by AS group_by,
COUNT(DISTINCT e.clinical_effective_date) AS visits
FROM cohort_gh2_encounter_raw e JOIN cohort_gh2 cr
ON e.person_id = cr.person_id
WHERE EXISTS (SELECT 'x' FROM gh2_f2fEncounters s
WHERE s.term = e.fhir_original_term)
AND e.clinical_effective_date IS NOT NULL
AND e.fhir_original_term IS NOT NULL
GROUP BY cr.group_by) b, (SELECT @row_no := 0) t;
SET @row_id = 0;
WHILE EXISTS (SELECT row_id from noVisitsTotal
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
UPDATE gh2_demographicsDataset d
JOIN noVisitsTotal nvt ON d.pseudo_id = nvt.group_by
SET d.F2fVisits_Total = nvt.visits
WHERE nvt.row_id > @row_id AND nvt.row_id <= @row_id + 1000;
SET @row_id = @row_id + 1000;
END WHILE;
Face to face visits are identified by the FHIR original terms in the Encounter table. The count distinct is used on the clinical effective date to identify the number of distinct visits. For example, add the total number of face to face visits in the last 1 year:
DROP TEMPORARY TABLE IF EXISTS lastyearvisits;
CREATE TEMPORARY TABLE lastyearvisits (
row_id INT,
group_by VARCHAR(255),
visits INT, PRIMARY KEY(row_id) ) AS
SELECT (@row_no := @row_no + 1) AS row_id,
b.group_by,
b.visits
FROM (SELECT cr.group_by AS group_by,
COUNT(DISTINCT e.clinical_effective_date) AS visits
FROM cohort_gh2_encounter_raw e JOIN cohort_gh2 cr
ON e.person_id = cr.person_id
WHERE EXISTS (SELECT 'x' FROM gh2_f2fEncounters s
WHERE s.term = e.fhir_original_term)
AND e.clinical_effective_date >
DATE_SUB(now(), INTERVAL 12 MONTH) -- last 1 yr
AND e.clinical_effective_date IS NOT NULL
AND e.fhir_original_term IS NOT NULL
GROUP BY cr.group_by) b, (SELECT @row_no := 0) t;
SET @row_id = 0;
WHILE EXISTS (SELECT row_id from lastyearvisits
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
UPDATE gh2_demographicsDataset d
JOIN lastyearvisits lyr ON d.pseudo_id = lyr.group_by
SET d.F2fVisits_1year = lyr.visits
WHERE lyr.row_id > @row_id AND lyr.row_id <= @row_id + 1000;
SET @row_id = @row_id + 1000;
END WHILE;
The clinical effective date is used to determine the time interval.
For example, add the total number of face to face visits for the last 5 years:
DROP TEMPORARY TABLE IF EXISTS lastfiveyearvisits;
CREATE TEMPORARY TABLE lastfiveyearvisits (
row_id INT,
group_by VARCHAR(255),
visits INT, PRIMARY KEY(row_id) ) AS
SELECT (@row_no := @row_no + 1) AS row_id,
b.group_by,
b.visits
FROM ( SELECT cr.group_by AS group_by,
COUNT(DISTINCT e.clinical_effective_date) AS visits
FROM cohort_gh2_encounter_raw e JOIN cohort_gh2 cr ON e.person_id =
cr.person_id
WHERE EXISTS (SELECT 'x' FROM gh2_f2fEncounters s
WHERE s.term = e.fhir_original_term)
AND e.clinical_effective_date > DATE_SUB(now(), INTERVAL 60 MONTH) --
last 5 yrs
AND e.clinical_effective_date IS NOT NULL
AND e.fhir_original_term IS NOT NULL
GROUP BY cr.group_by) b, (SELECT @row_no := 0) t;
SET @row_id = 0;
WHILE EXISTS (SELECT row_id from lastfiveyearvisits
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
UPDATE gh2_demographicsDataset d
JOIN lastfiveyearvisits lfyr ON d.pseudo_id = lfyr.group_by
SET d.F2fVisits_5years = lfyr.visits
WHERE lfyr.row_id > @row_id AND lfyr.row_id <= @row_id + 1000;
SET @row_id = @row_id + 1000;
END WHILE;
Note: the patient's name, age, and date of birth are embedded in the pseudo identifier string that needs to be de-anonymised before it is sent to the recipient.
Patient observations dataset
A typical patient observations dataset includes the following information:
A typical dataset can contain 100 to 200 fields, and usually there are several datasets to deliver.
Create table script for a patient diagnosis dataset table
In the following example the create table script produces the patient diagnoses dataset table:
DROP TABLE IF EXISTS gh2_diagnoses2dataset;
CREATE TABLE gh2_diagnoses2dataset (
Pseudo_id VARCHAR(255) NULL,
Pseudo_NHSNumber VARCHAR(255) NULL,
AsthmaECode VARCHAR(50) NULL,
AsthmaETerm VARCHAR(200) NULL,
AsthmaEDate VARCHAR(50) NULL,
AsthmaEmergeECode VARCHAR(50) NULL,
AsthmaEmergeETerm VARCHAR(200) NULL,
AsthmaEmergeEDate VARCHAR(50) NULL,
AsthmaResolvedECode VARCHAR(50) NULL,
AsthmaResolvedETerm VARCHAR(200) NULL,
AsthmaResolvedEDate VARCHAR(50) NULL,
COPDECode VARCHAR(50) NULL,
COPDETerm VARCHAR(200) NULL,
COPDEDate VARCHAR(50) NULL,
PulmonaryFibrosisECode VARCHAR(50) NULL,
PulmonaryFibrosisETerm VARCHAR(200) NULL,
PulmonaryFibrosisEDate VARCHAR(50) NULL,
InterstitialLungDiseaseECode VARCHAR(50) NULL,
InterstitialLungDiseaseETerm VARCHAR(200) NULL,
InterstitialLungDiseaseEDate VARCHAR(50) NULL,
AgeRelatedMuscularDegenerationECode VARCHAR(50) NULL,
AgeRelatedMuscularDegenerationETerm VARCHAR(200) NULL,
AgeRelatedMuscularDegenerationEDate VARCHAR(50) NULL,
GlaucomaECode VARCHAR(50) NULL,
GlaucomaETerm VARCHAR(200) NULL,
GlaucomaEDate VARCHAR(50) NULL,
RheumatoidArthritisECode VARCHAR(50) NULL,
RheumatoidArthritisETerm VARCHAR(200) NULL,
RheumatoidArthritisEDate VARCHAR(50) NULL,
SystemicLupusECode VARCHAR(50) NULL,
SystemicLupusETerm VARCHAR(200) NULL,
SystemicLupusEDate VARCHAR(50) NULL,
InflammatoryBowelDiseaseECode VARCHAR(50) NULL,
InflammatoryBowelDiseaseETerm VARCHAR(200) NULL,
InflammatoryBowelDiseaseEDate VARCHAR(50) NULL,
CrohnsDiseaseECode VARCHAR(50) NULL,
CrohnsDiseaseETerm VARCHAR(200) NULL,
CrohnsDiseaseEDate VARCHAR(50) NULL,
UlcerativeColitisCodeECode VARCHAR(50) NULL,
UlcerativeColitisCodeETerm VARCHAR(200) NULL,
UlcerativeColitisCodeEDate VARCHAR(50) NULL,
AtopicDermatitisECode VARCHAR(50) NULL,
AtopicDermatitisETerm VARCHAR(200) NULL,
AtopicDermatitisEDate VARCHAR(50) NULL,
InheritedMucociliaryClearanceECode VARCHAR(50) NULL,
InheritedMucociliaryClearanceETerm VARCHAR(200) NULL,
InheritedMucociliaryClearanceEDate VARCHAR(50) NULL,
PrimaryCiliaryDyskinesiaECode VARCHAR(50) NULL,
PrimaryCiliaryDyskinesiaETerm VARCHAR(200) NULL,
PrimaryCiliaryDyskinesiaEDate VARCHAR(50) NULL,
MelanomaECode VARCHAR(50) NULL,
MelanomaETerm VARCHAR(200) NULL,
MelanomaEDate VARCHAR(50) NULL,
ProstateCancerECode VARCHAR(50) NULL,
ProstateCancerETerm VARCHAR(200) NULL,
ProstateCancerEDate VARCHAR(50) NULL,
LungCancerECode VARCHAR(50) NULL,
LungCancerETerm VARCHAR(200) NULL,
LungCancerEDate VARCHAR(50) NULL,
SmallBowelCancerECode VARCHAR(50) NULL,
SmallBowelCancerETerm VARCHAR(200) NULL,
SmallBowelCancerEDate VARCHAR(50) NULL,
ColorectalCancerECode VARCHAR(50) NULL,
ColorectalCancerETerm VARCHAR(200) NULL,
ColorectalCancerEDate VARCHAR(50) NULL,
BreastCancerECode VARCHAR(50) NULL,
BreastCancerETerm VARCHAR(200) NULL,
BreastCancerEDate VARCHAR(50) NULL,
MiscarriageECode VARCHAR(50) NULL,
MiscarriageETerm VARCHAR(200) NULL,
MiscarriageEDate VARCHAR(50) NULL
);
ALTER TABLE gh2_diagnoses2dataset ADD INDEX gh2d2_pseudoid_idx (pseudo_id);
INSERT INTO gh2_diagnoses2dataset (pseudo_id) SELECT DISTINCT group_by FROM
cohort_gh2;
Each column of the dataset is named by combining the observational type, the time frame for the analysis, and the requested field type:
The dataset is populated with the pseudo identifiers from the patient cohort, and then SQL update queries populate the dataset table by calling multiple stored procedures.
Patient medications dataset
A typical patient medications dataset includes the following information:
Create table script for a patient medications dataset table
In the following example the create table script produces the patient medications dataset table:
CREATE TABLE bp2_medications (
pseudo_id VARCHAR(255) DEFAULT NULL,
nhsnumber VARCHAR(10) DEFAULT NULL,
codedate VARCHAR(20) DEFAULT NULL,
codeterm VARCHAR(255) DEFAULT NULL,
code VARCHAR(100) DEFAULT NULL,
codevalue VARCHAR(100) DEFAULT NULL,
codeunit VARCHAR(100) DEFAULT NULL
);
The dataset is populated with by calling a series of stored procedures.
Calling stored procedures
Once the datasets and cohorts have been generated, the next step is to populate the datasets using stored procedures.
A series of populate stored procedures call on other stored procedures to complete the update process:
Calling the populate stored procedures
In the following example the SQL script calls the populated stored procedures:
CALL populateCodeDateV2(0, 'CoronaryArteryDiseaseE', 'gh2_diagnoses1adataset', 0, '53741008,67682002,398274000,233970002,414545008,63739005', null, '703356002', null,'N');
CALL populateCodeDateV2(1, 'CoronaryArteryDiseaseL', 'gh2_diagnoses1adataset', 0, '53741008,67682002,398274000,233970002,414545008,63739005', null, '703356002', null,'N');
CALL populateCodeDateV2(0, 'AnginaE', 'gh2_diagnoses1adataset', 0, '194828000', null, null, null,'N');
CALL populateCodeDateV2(0, 'MyocardialInfarctionE', 'gh2_diagnoses1adataset', 0, '22298006,399211009', null, null, null,'N');
CALL populateCodeDateV2(0, 'CoronaryAngioplastyE', 'gh2_diagnoses1adataset', 0, '41339005', null, null, null,'N');
CALL populateCodeDateV2(0, 'CABGE', 'gh2_diagnoses1adataset', 0, '232717009,399261000', null, null, null,'N');
CALL populateCodeDateV2(0, 'AtrialFibrillationE', 'gh2_diagnoses1adataset', 0, '232717009', null, null, null,'N');
CALL populateCodeDateV2(1, 'AtrialFibrillationResolvedL', 'gh2_diagnoses1adataset', 0, '196371000000102', null, null, null,'N');
CALL populateCodeDateV2(0, 'AtrialFlutterE', 'gh2_diagnoses1adataset', 0, '5370000', null, null, null,'N');
CALL populateCodeDateV2(0, 'HeartFailureE', 'gh2_diagnoses1adataset', 0, '84114007,421518007,407596008,407597004,3545003,371037005', null, null, null,'N');
CALL populateCodeDateV2(0, 'TIAE', 'gh2_diagnoses1adataset', 0, '266257000,161511000', null, null, null,'N');
The procedure accepts a list of input parameters:
| Parameter | Data type | Description |
|---|---|---|
| filterType | INT | 1 latest, 0 earliest, 2 ever, 3 pivot |
| col | VARCHAR(100) the root of the column name | the root of the column name |
| datasetTable | VARCHAR(100) | table name of dataset |
| reset BIT 1 | BIT | 1 reset, 0 no reset of the dataset table |
| codesToAdd1 | VARCHAR(5000) | include all snomed parents and their children |
| codesToAdd2 | VARCHAR(5000) | include snomed parents only |
| codesToRemove3 | VARCHAR(5000) | exclude snomed parents and their children |
| codesToRemove4 | VARCHAR(5000) | exclude only snomed parents |
| ignorenulls | VARCHAR(1) | Y or N - whether to ignore null result value |
A typical patient medications dataset includes the following information:
USE data_extracts;
DROP PROCEDURE IF EXISTS populateCodeDateV2;
DELIMITER //
CREATE PROCEDURE populateCodeDateV2 (
IN filterType INT, -- 1 latest, 0 earliest, 2 ever, 3
pivot
IN col VARCHAR(100), -- the root of the column name
IN datasetTable VARCHAR(100), -- table name of dataset
IN reset BIT, -- 1 reset, 0 no reset
IN codesToAdd1 VARCHAR(5000), -- all parents and their children
IN codesToAdd2 VARCHAR(5000), -- parents only
IN codesToRemove3 VARCHAR(5000), -- parents and their children to be excluded
IN codesToRemove4 VARCHAR(5000), -- just parents to be excluded
IN ignorenulls VARCHAR(1)
)
BEGIN
-- clear out gh2_snomeds table
DELETE FROM gh2_snomeds WHERE cat_id IN (1, 2, 3, 4);
-- clear out gh2_store table
DELETE FROM gh2_store WHERE id IN (1, 2, 3, 4);
-- get snomeds
IF codesToAdd1 IS NOT NULL THEN
CALL storeSnomedString (codesToAdd1, 1);
CALL getAllSnomedsFromSnomedString (1);
END IF;
IF codesToAdd2 IS NOT NULL THEN
CALL storeSnomedString (codesToAdd2, 2);
CALL getAllSnomedsFromSnomedString (2);
END IF;
IF codesToRemove3 IS NOT NULL THEN
Retrieving SNOMED codes
Fig 6.2 Shows how the SNOMED codes are being retrieved using stored procedures
CALL storeSnomedString (codesToRemove3, 3);
CALL getAllSnomedsFromSnomedString (3);
END IF;
IF codesToRemove4 IS NOT NULL THEN
CALL storeSnomedString (codesToRemove4, 4);
CALL getAllSnomedsFromSnomedString (4);
END IF;
CALL filterObservationsV2(filterType,1,ignorenulls);
-- reset columns
IF (reset = 1) THEN
SET @reset_sql = CONCAT('UPDATE ', datasetTable, ' SET ',
col, "Code = null, ",
col, "Term = null, ",
col, "Date = null");
PREPARE resetStmt FROM @reset_sql;
EXECUTE resetStmt;
DEALLOCATE PREPARE resetStmt;
END IF;
DROP TEMPORARY TABLE IF EXISTS qry_tmp;
CREATE TEMPORARY TABLE qry_tmp (
row_id INT,
group_by VARCHAR(255),
original_code VARCHAR(20),
original_term VARCHAR(200),
clinical_effective_date DATE, PRIMARY KEY(row_id)
) AS
SELECT (@row_no := @row_no+1) AS row_id,
f.group_by,
f.original_code,
f.original_term,
f.clinical_effective_date
FROM filteredObservationsV2 f, (SELECT @row_no := 0) t;
SET @row_id = 0;
WHILE EXISTS (SELECT row_id from qry_tmp
WHERE row_id > @row_id AND row_id <= @row_id + 1000) DO
SET @sql = CONCAT('UPDATE ', datasetTable,
' d JOIN qry_tmp f ON d.pseudo_id = f.group_by SET ',
col, "Code = f.original_code, ",
col, "Term = f.original_term, ",
col, "Date = date_format(f.clinical_effective_date,
'%d/%m/%Y')
WHERE f.row_id > @row_id AND f.row_id <= @row_id + 1000");
PREPARE stmt FROM @sql;
EXECUTE stmt;
DEALLOCATE PREPARE stmt;
SET @row_id = @row_id + 1000;
END WHILE;
END//
DELIMITER ;
Retrieving SNOMED codes
The following diagram shows how the SNOMED codes are being retrieved using stored procedures:
Calling the storeSnomedString procedure
The first step of the procedure is to find all the SNOMED codes given a set of parent codes from the input parameter. We pass the codes into the procedure as a comma separated list.
We call the storeSnomedString procedure to separate the list of SNOMED codes into their individual values and store them into a temporary table:
USE data_extracts;
DROP PROCEDURE IF EXISTS storeSnomedString;
DELIMITER //
CREATE PROCEDURE storeSnomedString (
IN stringValue VARCHAR(5000),
IN cat_id INT
)
BEGIN
DECLARE front VARCHAR(5000) DEFAULT NULL;
DECLARE frontlen INT DEFAULT NULL;
DECLARE TempValue VARCHAR(5000) DEFAULT NULL;
iterator:
LOOP
IF LENGTH(TRIM(stringValue)) = 0 OR stringValue IS NULL THEN
LEAVE iterator;
END IF;
SET front = SUBSTRING_INDEX(stringValue, ',', 1);
SET frontlen = LENGTH(front);
SET TempValue = TRIM(front);
INSERT INTO gh2_store (id, org_snomed_id)
VALUES (cat_id, CAST(TempValue AS SIGNED));
SET stringValue = INSERT(stringValue, 1, frontlen + 1, '');
END LOOP;
END//
DELIMITER ;
DROP TABLE IF EXISTS gh2_store;
CREATE TABLE gh2_store (
id INT,
org_snomed_id BIGINT
);
ALTER TABLE gh2_store ADD INDEX gh2_store_idx (org_snomed_id);
We repeat this process for each CAT ID flag passed.
Calling the getAllSnomedsFromSnomedString procedure
We call another procedure getAllSnomedsFromSnomedString to loop through the temporary table, and depending on the CAT ID flag being passed, we retrieve all the corresponding child codes.
USE data_extracts;
DROP PROCEDURE IF EXISTS getAllSnomedsFromSnomedString;
DELIMITER //
CREATE PROCEDURE getAllSnomedsFromSnomedString (p_cat_id INT)
BEGIN
DECLARE done INT;
DECLARE l_parent_id BIGINT;
DECLARE c_get_snomeds CURSOR FOR SELECT org_snomed_id
FROM gh2_store WHERE id = p_cat_id ;
DECLARE CONTINUE HANDLER FOR NOT FOUND SET done = 1;
SET done = 0;
OPEN c_get_snomeds;
iterator:
WHILE (done = 0) DO
FETCH c_get_snomeds INTO l_parent_id;
IF done = 1 THEN
LEAVE iterator;
END IF;
IF p_cat_id IN (1, 3) THEN
INSERT INTO gh2_snomeds (snomed_id, cat_id)
SELECT DISTINCT
l_parent_id AS snomed_id,
p_cat_id AS cat_id
UNION
SELECT s.subtypeid AS snomed_id,
p_cat_id AS cat_id
FROM rf2.sct2_transitiveclosure s
WHERE s.supertypeid = l_parent_id
AND s.active = 1
UNION
SELECT s.subtypeid AS snomed_id,
p_cat_id AS cat_id
FROM rf2.sct2_transitiveclosure s
WHERE s.supertypeid IN (SELECT s1.subtypeid
FROM rf2.sct2_transitiveclosure s1
WHERE s1.supertypeid = l_parent_id)
AND s.active = 1;
ELSE
INSERT INTO gh2_snomeds (snomed_id, cat_id)
SELECT l_parent_id AS snomed_id,
p_cat_id AS cat_id;
END IF;
END WHILE iterator;
CLOSE c_get_snomeds;
SET done = 0;
END //
DELIMITER ;
The resultant set of codes is stored in a temporary table that is used to query observational data.
DROP TABLE IF EXISTS gh2_snomeds;
CREATE TABLE gh2_snomeds (
cat_id INT,
snomed_id BIGINT
);
ALTER TABLE gh2_snomeds ADD INDEX gh2_cat_idx (cat_id);
ALTER TABLE gh2_snomeds ADD INDEX gh2_sno_idx (snomed_id);
The following diagram shows the transitive closure table for looking up the hierarchical information of the SNOMED code:
 Find the corresponding child codes in the subtypeid column by matching the parent code on the supertypeid column.
Find the corresponding child codes in the subtypeid column by matching the parent code on the supertypeid column.
Retrieving filtered Observations
Using the prepared observations cohort, the next step is to filter out this cohort based on the SNOMED codes list and the timeframe, which have been passed in as input parameters from the populate procedure.
The following diagram shows the processing of the FilterObservations procedure:
 Calling FilterObservations Procedure
Calling FilterObservations Procedure
| Parameter | Data type | Description |
|---|---|---|
| filterType | INT | 0 earliest, 1 latest, 2 ever, 3 pivot around cohort pivot date (6 months), 4 all Since |
| toCreate | INT | 1 is the default value - to call createObservationsFromCohort |
| ignorenulls | VARCHAR(1) | Y or N - whether to ignore null result value |
USE data_extracts;
DROP PROCEDURE IF EXISTS filterObservationsV2;
DELIMITER //
CREATE PROCEDURE filterObservationsV2 (
IN filterType INT,
-- 0 earliest, 1 latest, 2 ever,
-- 3 pivot around cohort pivot date (6 months),
-- 4 allSince
IN toCreate INT, -- 1 to createObservationsFromCohortv2
IN ignorenulls VARCHAR(1) -- Y or N
)
BEGIN
IF (toCreate = 1) THEN
CALL createObservationsFromCohortv2 (filterType);
END IF;
DROP TABLE IF EXISTS filteredObservationsV2;
IF (filterType = 0) THEN -- earliest
IF (ignorenulls = 'Y') THEN
CREATE TABLE filteredObservationsV2 as
SELECT
ob.id,
ob.group_by,
ob.patient_id,
ob.person_id,
ob.original_code,
ob.original_term,
ob.result_value,
ob.clinical_effective_date,
ob.result_value_units,
ob.age_years,
ob.rnk
FROM (
SELECT o.id,
o.group_by,
o.patient_id,
o.person_id,
o.original_code,
o.original_term,
o.result_value,
o.clinical_effective_date,
o.result_value_units,
o.age_years,
@currank := IF(@curperson = o.person_id,
@currank + 1, 1) AS rnk,
@curperson := o.person_id AS cur_person
FROM observationsFromCohortV2 o,
(SELECT @currank := 0, @curperson := 0) r
WHERE o.result_value IS NOT NULL
ORDER BY o.person_id, o.clinical_effective_date ASC,
o.id ASC -- earliest
) ob
WHERE ob.rnk = 1;
ELSE
CREATE TABLE filteredObservationsV2 as
SELECT
ob.id,
ob.group_by,
ob.patient_id,
ob.person_id,
ob.original_code,
ob.original_term,
ob.result_value,
ob.clinical_effective_date,
ob.result_value_units,
ob.age_years,
ob.rnk
FROM (
SELECT o.id,
o.group_by,
o.patient_id,
o.person_id,
o.original_code,
o.original_term,
o.result_value,
o.clinical_effective_date,
o.result_value_units,
o.age_years,
@currank := IF(@curperson = o.person_id,
@currank + 1, 1) AS rnk,
@curperson := o.person_id AS cur_person
FROM observationsFromCohortV2 o,
(SELECT @currank := 0, @curperson := 0) r
ORDER BY o.person_id, o.clinical_effective_date ASC,
o.id ASC -- earliest
) ob
WHERE ob.rnk = 1;
END IF;
ELSEIF (filterType = 2 OR filterType = 4) THEN -- ever or allSince
IF (ignorenulls = 'Y') THEN
CREATE TABLE filteredObservationsV2 AS
SELECT DISTINCT
-- mc.id,
mc.group_by,
mc.patient_id,
mc.person_id,
mc.original_code,
mc.original_term,
mc.result_value,
mc.clinical_effective_date,
mc.result_value_units,
mc.age_years
FROM observationsFromCohortV2 mc
WHERE mc.result_value IS NOT NULL;
ELSE
CREATE TABLE filteredObservationsV2 AS
SELECT DISTINCT
-- mc.id,
mc.group_by,
mc.patient_id,
mc.person_id,
mc.original_code,
mc.original_term,
mc.result_value,
mc.clinical_effective_date,
mc.result_value_units,
mc.age_years
FROM observationsFromCohortV2 mc;
END IF;
ELSEIF (filterType = 1 or filterType = 3) THEN -- latest or pivot
IF (ignorenulls = 'Y') THEN
CREATE TABLE filteredObservationsV2 as
SELECT
ob.id,
ob.group_by,
ob.patient_id,
ob.person_id,
ob.original_code,
ob.original_term,
ob.result_value,
ob.clinical_effective_date,
ob.result_value_units,
ob.age_years,
ob.rnk
FROM (
SELECT o.id,
o.group_by,
o.patient_id,
o.person_id,
o.original_code,
o.original_term,
o.result_value,
o.clinical_effective_date,
o.result_value_units,
o.age_years,
@currank := IF(@curperson = o.person_id,
@currank + 1, 1) AS rnk,
@curperson := o.person_id AS cur_person
FROM observationsFromCohortV2 o,
(SELECT @currank := 0, @curperson := 0) r
WHERE o.result_value IS NOT NULL
ORDER BY o.person_id, o.clinical_effective_date DESC,
o.id DESC -- latest
) ob
WHERE ob.rnk = 1;
ELSE
CREATE TABLE filteredObservationsV2 as
SELECT
ob.id,
ob.group_by,
ob.patient_id,
ob.person_id,
ob.original_code,
ob.original_term,
ob.result_value,
ob.clinical_effective_date,
ob.result_value_units,
ob.age_years,
ob.rnk
FROM (
SELECT o.id,
o.group_by,
o.patient_id,
o.person_id,
o.original_code,
o.original_term,
o.result_value,
o.clinical_effective_date,
o.result_value_units,
o.age_years,
@currank := IF(@curperson = o.person_id,
@currank + 1, 1) AS rnk,
@curperson := o.person_id AS cur_person
FROM observationsFromCohortV2 o,
(SELECT @currank := 0, @curperson := 0) r
ORDER BY o.person_id, o.clinical_effective_date DESC,
o.id DESC -- latest
) ob
WHERE ob.rnk = 1;
END IF;
ELSE
SIGNAL SQLSTATE '45000'
SET MESSAGE_TEXT = 'filterType not recognised';
END IF;
END//
DELIMITER ;
Calling the createobservationsFromCohort procedure
The createobservationsFromCohort procedure is called from the FilterObservations procedure; the filter type is passed as the input parameter:
call createObservationsFromCohortv2 (filterType);
USE data_extracts;
DROP PROCEDURE IF EXISTS createObservationsFromCohortV2;
DELIMITER //
CREATE PROCEDURE createObservationsFromCohortV2 (
IN filterType INT
-- 0 earliest, 1 latest, 2 ever, 3 pivot around cohort_gh2 pivot date (6
months)
)
BEGIN
DROP TEMPORARY TABLE IF EXISTS gh2_snomeds_tmp;
CREATE TEMPORARY TABLE gh2_snomeds_tmp AS
SELECT cat_id,
snomed_id
FROM gh2_snomeds
WHERE cat_id IN (3, 4);
DELETE t1 FROM gh2_snomeds t1 JOIN gh2_snomeds_tmp t2
ON t1.snomed_id = t2.snomed_id
WHERE t1.cat_id IN (1, 2);
DROP TABLE IF EXISTS observationsFromCohortV2;
IF (filterType = 3) THEN
-- pivot over 6 months from pivot date (already set in cohort)
CREATE TABLE observationsFromCohortV2 AS
SELECT DISTINCT
o.id,
o.patient_id,
o.person_id,
o.group_by,
o.clinical_effective_date,
o.original_code,
SUBSTRING(o.original_term, 1, 200) AS original_term,
o.result_value,
o.result_value_units,
o.age_years
FROM cohort_gh2_observations o JOIN cohort_gh2 c
ON o.group_by = c.group_by
WHERE EXISTS (SELECT 'x' FROM gh2_snomeds s
WHERE s.cat_id IN (1, 2)
AND s.snomed_id = o.original_code)
AND o.clinical_effective_date IS NOT NULL
AND o.clinical_effective_date
BETWEEN DATE_SUB(c.pivot_date, INTERVAL 6 MONTH)
AND DATE_SUB(c.pivot_date, INTERVAL -6 MONTH);
ELSE
-- latest or earliest or ever (so all observations)
CREATE TABLE observationsFromCohortV2 AS
SELECT DISTINCT
o.id,
o.patient_id,
o.person_id,
o.group_by,
o.clinical_effective_date,
o.original_code,
SUBSTRING(o.original_term, 1, 200) AS original_term,
o.result_value,
o.result_value_units,
o.age_years
FROM cohort_gh2_observations o
WHERE EXISTS (SELECT 'x' FROM gh2_snomeds s
WHERE s.cat_id IN (1, 2)
AND s.snomed_id = o.original_code)
AND o.clinical_effective_date IS NOT NULL;
END IF;
ALTER TABLE observationsFromCohortV2 ADD INDEX gh2_obv_pat_idx (patient_id);
END//
DELIMITER ;